Biochemistry 2/e - Garrett & Grisham
Chapter 32
The Genetic Code
to accompany
Biochemistry, 2/e
by
Reginald Garrett and Charles Grisham
All rights reserved. Requests for permission to make copies of any part of the work
should be mailed to: Permissions Department, Harcourt Brace & Company,
6277
Sea Harbor Drive, Orlando, Florida 32887-6777
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Outline
•
•
•
•
32.1 Elucidating the Genetic Code
32.2 The Nature of the Genetic Code
32.3 The Second Genetic Code
32.4 Codon-Anticodon Pairing, Third-Base
Degeneracy and the Wobble Hypothesis
• 32.5 Codon Usage
• 32.6 Nonsense Suppression
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Translating the Message
How does the sequence of mRNA translate into the
sequence of a protein?
• What is the genetic code?
• How do you translate the "four-letter code" of
mRNA into the "20-letter code" of proteins?
• And what are the mechanics like? There is no
obvious chemical affinity between the purine and
pyrimidine bases and the amino acids that make
protein.
• As a "way out" of this dilemma, Crick proposed
"adapter molecules" - they are tRNAs!
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
The Collinearity of Gene and
Protein Structures
• Watson and Crick's structure for DNA, together with
Sanger's demonstration that protein sequences
were unique and specific, made it seem likely that
DNA sequence specified protein sequence
• Yanofsky provided better evidence in 1964: he
showed that the relative distances between
mutations in DNA were proportional to the distances
between amino acid sunstitutions in E. coli
tryptophan synthase
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Elucidating the Genetic Code
• A triplet code is required: 43 = 64, but
42 = 16 - not enough for 20 amino
acids
• But is the code overlapping?
• See Figure 32.2
• And is the code punctuated?
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
The Nature of the Genetic
Code
• A group of three bases codes for one
amino acid
• The code is not overlapping
• The base sequence is read from a fixed
starting point, with no punctuation
• The code is degenerate (in most cases,
each amino acid can be designated by
any of several triplets
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Biochemists Break the Code
•
•
•
•
•
Assignment of "codons" to their respective amino
acids was achieved by in vitro biochemistry
Marshall Nirenberg and Heinrich Matthaei showed
that poly-U produced polyphenylalanine in a cellfree solution from E. coli
Poly-A gave polylysine
Poly-C gave polyproline
Poly-G gave polyglycine
But what of others?
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Getting at the Rest of the Code
• Work with nucleotide copolymers (poly (A,C), etc.),
revealed some of the codes
• But Marshall Nirenberg and Philip Leder cracked
the entire code in 1964
• They showed that trinucleotides bound to
ribosomes could direct the binding of specific
aminoacyl-tRNAs (See Figure 31.6)
• By using C-14 labelled amino acids with all the
possible trinucleotide codes, they elucidated all 64
correspondences in the code (Table 32.3)
• Read also about Khorana's experiment
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Features of the Genetic Code
• All the codons have meaning: 61 specify amino
acids, and the other 3 are "nonsense" or "stop"
codons
• The code is unambiguous - only one amino acid is
indicated by each of the 61 codons
• The code is degenerate - except for Trp and Met,
each amino acid is coded by two or more codons
• Codons representing the same or similar amino
acids are similar in sequence
• 2nd base pyrimidine: usually nonpolar amino acid
• 2nd base purine: usually polar or charged aa
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
AA Activation for Prot. Synth.
•
•
•
•
•
The Aminoacyl-tRNA Synthetases
Codons are recognized by aminoacyl-tRNAs
Base pairing must allow the tRNA to bring its
particular amino acid to the ribosome
But aminoacyl-tRNAs do something else:
activate the amino acid for transfer to peptide
Aminoacyl-tRNA synthetases do the critical job linking the right amino acid with "cognate" tRNA
Two levels of specificity - one in forming the
aminoacyl adenylate and one in linking to tRNA
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Aminoacyl-tRNA Synthetases
•
•
•
•
Mechanism and specificity
Deacylase activity "edits" and hydrolyzes
misacylated aminoacyl-tRNAs
Despite common function, the synthetases are
a diverse collection of enzymes
Four different quaternary structures: , 2, 4
and 22
Subunits from 334 to more than 1000 residues
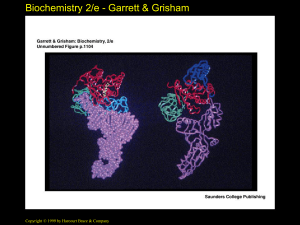
• Two different mechanisms (See Figure 32.5)
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Recognition of tRNAs
by the aminoacyl-tRNA synthetases
• Anticodon region is not the only
recognition site
• The "inside of the L" and other regions
of the tRNA molecule are also important
• Read pages 1080-1082 on specificity of
several aminoacyl-tRNA synthetases
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Third-Base Degeneracy
and the Wobble Hypothesis
• Codon-anticodon pairing is the crucial feature of
the "reading of the code"
• But what accounts for "degeneracy": are there 61
different anticodons, or can you get by with fewer
than 61, due to lack of specificity at the third
position?
• Crick's Wobble Hypothesis argues for the second
possibility - the first base of the anticodon (which
matches the 3rd base of the codon) is referred to
as the "wobble position"
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
Copyright © 1999 by Harcourt Brace & Company
Biochemistry 2/e - Garrett & Grisham
The Wobble Hypothesis
• The first two bases of the codon make normal
(canonical) H-bond pairs with the 2nd and 3rd
bases of the anticodon
• At the remaining position, less stringent rules apply
and non-canonical pairing may occur
• The rules: first base U can recognize A or G, first
base G can recognize U or C, and first base I can
recognize U, C or A (I comes from deamination of
A)
• Advantage of wobble: dissociation of tRNA from
mRNA is faster and protein synthesis too
Copyright © 1999 by Harcourt Brace & Company