Research Article
15
Determination of apoptosis, proliferation status
and O6-methylguanine DNA methyltransferase
methylation profiles in different immunophenotypic
profiles of diffuse large B-cell lymphoma
Diffüz büyük B-hücreli lenfomanın farklı immünofenotipik
profillerinde apoptozis, proliferasyon durumu ve O6-metilguanin DNA
metiltransferaz metilasyon profillerinin tespiti
Nilay Şen Türk1, Nazan Özsan2, Vildan Caner3, Nedim Karagenç3, Füsun Düzcan4,
Ender Düzcan1, Mine Hekimgil2
1Department
of Pathology, Faculty of Medicine, Pamukkale University, Denizli, Turkey
of Pathology, Faculty of Medicine, Ege University, İzmir, Turkey
3Department of Medical Biology, Faculty of Medicine, Pamukkale University, Denizli, Turkey
4Department of Medical Genetics, Faculty of Medicine, Pamukkale University, Denizli, Turkey
2Department
Abstract
Objective: Our aim was to investigate the expression of apoptosis-associated proteins (bcl-2, bcl-xl,
bax, bak, bid), apoptotic index (AI) and proliferation index (PI) in germinal center B-cell-like immunophenotypic profile (GCB) and non-GCB of diffuse large B-cell lymphoma (DLBCL).
Materials and Methods: The methylation status of the promoter region of O6-methylguanine-DNA
yerine O6-methylguanine-DNA methyltransferase (MGMT) gene and its relation with immunophenotypic differentiation of DLBCLs were also investigated. 101 cases were classified as GCB (29 cases) or
non-GCB (72 cases). Apoptosis-associated proteins and PI were determined by IHC, and TUNEL
method was used to determine AI. MGMT methylation analysis was performed by real-time PCR.
Results: The PI was significantly higher in GCB compared with non-GCB (p=0.011). Percentage of
cells stained with bcl-6 was positively correlated with the percentage of cells expressing bcl-2
(p=0.023), AI (p=0.006) and PI (p<0.001), while a significant negative correlation was observed
with the percentage of cells expressing bax (p=0.027). The percentage of cells stained with MUM1
showed a significantly positive correlation with the percentage of cells expressing bcl-xl (p=0.003), bid
(p=0.002), AI (p<0.001), and PI (p=0.001). MGMT methylation analysis was performed in 95
samples, and methylated profile was found in 31 cases (32.6%). GCB was found in 6 cases (22.2%)
and non-GCB was determined in 25 cases (36.8%) out of 31 with MGMT methylated samples. There
was no significant association between MGMT methylation status and immunophenotypic profiles
(p=0.173).
Address for Correspondence: Asst. Prof. Nilay Şen Türk, Department of Pathology, Faculty of Medicine, Pamukkale University, Denizli, Turkey
Phone: +90 258 361 39 16 E-mail: [email protected]
doi:10.5152/tjh.2010.37
16
Türk et al.
Apoptosis, proliferation in large B-cell lymphoma
Turk J Hematol 2011; 28: 15-26
Conclusion: These results suggest that bcl-6 protein expression may be responsible for the high PI in
GCB. Additionally, we found that apoptosis-associated proteins were not significantly associated with
immunophenotypic profiles. (Turk J Hematol 2011; 28: 15-26)
Key words: Lymphoma, B-cell, apoptosis, proliferation, methylation
Received: March 1, 2010
Accepted: June 29, 2010
Özet
Amaç: Diffüz büyük B-hücreli lenfoma (DBBHL)’nın germinal merkez B-hücresi benzeri (GCB) ve
non-GCB profillerinde apoptozis-ilişkili proteinlerin (bcl-2, bcl-xl, bax, bak, bid) ekspresyonunu, apoptotic indeksi (AI) ve proliferasyon indeksi (PI)’ni araştırmaktır. Ayrıca, O6-methylguanine-DNA methyltransferase (MGMT) geninin promoter bölgesinin metilasyon durumunu ve onun DBBHL’nın
immünofenotipik diferansiyasyonuyla ilişkisini araştırmaktır.
Yöntem ve Gereçler: 101 olgu GCB (29 olgu) ve non-GCB (72 olgu) olarak sınıflandırıldı. Apoptozisilişkili proteinler ve PI immünohistokimyasal olarak saptandı ve TUNEL yöntemi AI’i belirlemek için
kullanıldı. MGMT metilasyon analizi, real-time PCR ile gerçekleştirildi.
Bulgular: PI, non-GCB ile karşılaştırıldığında GCB’de anlamlı şekilde yüksek saptandı (p=0.011).
Bcl-6 ile p: PI, non-GCB ile karşılaştırıldığında GCB’de anlamlı şekilde yüksek saptandı (p=0.011).
Bcl-6 ile pozitif boyanan hücrelerin yüzdesi bcl-2 (p=0.023), AI (p=0.006), ve PI (p<0.001) eksprese
eden hücrelerin yüzdesi ile pozitif şekilde korele iken, bax eksprese eden hücrelerin yüzdesi ile negatif korelasyon gözlendi (p=0.027). MUM1 ile boyanan hücrelerin yüzdesi bcl-xl (p=0.003), bid
(p=0.002), AI (p<0.001) ve PI (p=0.001) eksprese eden hücrelerin yüzdesi ile anlamlı şekilde pozitif
korelasyon gösterdi. MGMT metilasyon analizi 95 örneğe uygulandı ve metilasyon profili 31 olguda
(%32.6) saptandı. 31 MGMT metile örnekten 6 olgunun (%22.2) GCB ve 25 olgunun (%36.8) nonGCB olduğu belirlendi. MGMT metilasyon durumu ve immünofenotipik profiler arasında anlamlı ilişki
saptanmadı (p=0.173).
Sonuç: Bu bulgular, bcl-6 protein ekspresyonunun GCB’de yüksek PI’inden sorumlu olabileceğini öne
sürmektedir. Ek olarak, apoptozis-ilişkili proteinlerin immünofenotipik profilerle anlamlı ilişki
göstermediğini saptadık. (Turk J Hematol 2011; 28: 15-26)
Anahtar kelimeler: Büyük B-hücreli lenfomalarda apoptosis ve proliferasyon
Geliş tarihi: 1 Mart 2010
Kabul tarihi: 29 Haziran 2010
Introduction
Diffuse large B-cell lymphoma (DLBCL) constitutes the largest group of aggressive lymphomas in
adults and 30-40% of adult non-Hodgkin lymphomas (NHL) in western countries [1,2]. DLBCLs
show diversity in clinical presentation, morphology
and genetic and molecular properties, which suggests that these tumors represent a heterogeneous
group of neoplasms rather than a single clinicopathologic entity [2,3]. Therefore, patients who are
diagnosed as DLBCL could show remarkably different history, clinical behavior and outcome [1].
Different mechanisms such as deregulation of
cell cycle and apoptotic pathways are involved in
the pathogenesis of DLBCL. As for the molecular
pathogenesis of DLBCL, distinct chromosomal
translocations and aberrant somatic hypermutations as well as numerical chromosomal anomalies
such as duplications and deletions, which are also
common to other malignancies, have been reported [4-7].
DLBCL originates from germinal center (GC) and
post-GC B-cells that normally have encountered
with antigen [8,9]. Different methods are applied to
determine B-cell differentiation antigens in DLBCL.
In spite of the distinct advantages of cDNA and oligonucleotide microarray techniques, immunohistochemistry is a commonly used method to determine
the GC B-cell-like (GCB) and non-GCB-like profiles
because it is cheap and easily applied [10-12]. Hans
et al. [11] reported that the classification of DLBCL
into GCB and non-GCB profiles based on CD10/
bcl-6/MUM1 immunophenotypic differentiation is
prognostically relevant to the cDNA classification in
71% of GCB and in 88% of non-GCB. It is well known
that the expression of bcl-6 and CD10 are associated with increased apoptosis and proliferation in
lymphoid malignancies [13-18]. Bai et al. [10]
reported that increased expression of apoptotic
index (AI) in DLBCL with GCB profile is associated
with high expression of the pro-apoptotic proteins
(bax, bak, bid) and low expression of the antiapoptotic protein (bcl-xl). However, data related to apop-
Turk J Hematol 2011; 28: 15-26
tosis and proliferation status in CD10/bcl-6/MUM1
immunophenotypic differentiation profiles is considerably limited. Although the immunohistochemical expression of the apoptosis-associated bcl-2
family proteins, bcl-2, bax, bak, and mcl1, was
reported in DLBCLs [19-28], the expression levels of
bcl-xl, bad, bid proteins and their relations with the
status of apoptosis and proliferation were not extensively analyzed in these lymphomas [25].
Altered DNA methylation profile has been comprehensively studied in the pathogenesis of cancer.
Compared with normal cells, cancer cells frequently demonstrate genome-wide hypomethylation,
hypermethylation of tumor suppressor gene, and
loss of genomic imprinting [29]. In human cancers,
the gene encoding the DNA-repair enzyme O6methylguanine-DNA methyltransferase (MGMT) is
not commonly mutated or deleted. Loss of MGMT
expression is mainly due to epigenetic changes,
specifically methylation of the promoter region
[1,30]. MGMT protects cells from the toxicity of environmental and therapeutic alkylating agents, which
frequently target the O6 position of guanine.
Inactivation of the MGMT gene via hypermethylation of its promoter region increases sensitivity of
cells to the genotoxic effect of alkylating agents
both in vitro and in vivo [1,3]. Recent research has
focused on the relationship between MGMTmethylation status and immunophenotypic differentiation in DLBCLs [30,31].
The aim of this study was to investigate the
expression profiles of apoptosis-associated proteins
(bcl-2, bcl-xl, bax, bak, bid), apoptotic index (AI),
and proliferation index (PI) in GCB and non-GCB
immunophenotypic profiles of DLBCL. In addition,
the methylation status of the promoter region of the
MGMT gene and its relation with immunophenotypic differentiation of DLBCLs were investigated.
Materials and Methods
Materials
A total of 101 cases of de novo DLBCLs, diagnosed according to the World Health Organization
(WHO) classification [2], were obtained from the
files of the Department of Pathology, Faculty of
Medicine, Ege University. Clinicopathological
parameters for all patients were obtained from the
pathology records. Ethical committee approval was
obtained for this study.
Türk et al.
Apoptosis, proliferation in large B-cell lymphoma
17
Immunohistochemistry
For immunohistochemical staining, sections of
5-μm-thickness were cut from formalin-fixed, paraffin-embedded tissue blocks and placed on electrostatic-charged, poly-L-lysine-coated slides (X-traTM,
Surgipath Medical Industries, Richmond, IL, USA).
Sections were dehydrated at 60ºC for a minimum of
2 hours (h). All immunostaining procedures including deparaffinization and antigen retrieval processes were performed on BenchMark XT® automated
stainer (Venatana Medical Systems, USA). After
counterstaining of the slides with hematoxylin in
automated stainer, dehydration, incubation in
xylene and mounting processes were performed
manually, and immunostaining procedure was
completed. Bcl-6 (dilution: 1/20, clone: P1F6, Dako
SA, Glostrup, Denmark), CD10 (dilution: 1/25, clone:
56C6, Spring Bioscience, Pleasanton, CA, USA),
IRF4/MUM1 (dilution: 1/25, clone: MUM1p, Dako SA,
Glostrup, Denmark), bcl-2 (dilution: 1/40, clone: bcl2/100/DS, Novocastra, Newcastle upon Tyne, UK),
bcl-xl (dilution: 1/20, clone: 2H12, Zymed, South San
Francisco, CA, USA), bax (dilution: 1/200, code:
A3533, Dako SA, Glostrup, Denmark), bak (dilution:
1/100, code: A3538, Dako SA, Glostrup, Denmark),
bid (dilution: 1/100, clone: NB110-40718, Novus,
Littleton, CO, USA), and Ki-67 (dilution: 1/150, clone:
MIB-1, Dako SA, Glostrup, Denmark) were used as
primary antibodies.
Reactive lymph nodes and normal thymic tissue
samples were used as positive controls. Negative
controls were treated with the same immunohistochemical method by omitting the primary antibody.
At least 10 fields selected on the basis that they
contained immunopositive cells were counted by
using the 40x objective lens on the light microscope.
The number of immunopositive cells was divided
by the total number of the counted cells, and the
expression was defined as the percentage of positive cells. CD10, bcl-6, and MUM1 proteins were
considered positive when at least 25% of neoplastic
cells were immunopositive according to the previously defined criteria [12]. The CD10/bcl-6/MUM1
immunophenotypes and their designation to GCB
and non-GCB profiles were determined according
to the classification by Hans et al. [11]. The expressions of bcl-2, bcl-xl, bax, bak, and bid proteins
were considered positive when at least 10% of neoplastic cells were immunopositive [10]. PI with
18
Türk et al.
Apoptosis, proliferation in large B-cell lymphoma
Ki-67 was determined as the percentage of positive
cells within the total number of the counted cells.
Tunel Method
The terminal deoxynucleotidyl-transferase (TdT)mediated in situ labeling technique (TUNEL; in situ
Cell Death Detection Kit, POD, Roche) was performed on the 5-μm-thick sections of formalinfixed, paraffin-embedded tissue for demonstration
of DNA fragmentation. Briefly, after deparaffinization and dehydration, slides were rinsed in phosphate-buffered saline (PBS) (pH 7.4). The peroxidase activity was blocked by incubation for 15 minutes (min) in 3% hydrogen peroxide in PBS at room
temperature. Tissue sections were digested by incubation for 30 min with proteinase K (20 μg/ml) at
37ºC, and then were rinsed in PBS. TUNEL reaction
mixture was prepared according to the manufacturer’s recommendations, and a mixture of 50 μL
per slide was added. Slides were incubated for 1 h
at 37ºC in dark. One positive control and two negative controls were included in each set of experiments. Reactive lymph nodes were used as positive
controls. Negative controls were treated similarly by
omitting the TdT reaction step. Slides, once again,
were rinsed in PBS. To examine the slides in light
microscope, 50 μl Converter-peroxidase (POD) was
added per slide and slides were incubated for 30
min at 37ºC in humid and dark conditions. Slides
were rinsed in PBS, and then were incubated with
3,3’-diaminobenzidine tetrahydrochloride (DAB
Substrate, Roche) for 10 min at room temperature.
Slides were counterstained with Harris’ hematoxylin and mounted. The evaluation of the results was
performed as Bai et al. [32] had described previously. Morphologically intact TUNEL-positive cells
were considered as positive and referred to as
apoptotic cells. The number of apoptotic cells was
recorded by using the 40X objective in at least 10
randomly selected fields. The AI was expressed as
a percentage of the number of apoptotic cells within the total number of counted cells.
O6- Methylguanine-DNA Methyltransferase
Methylation Analysis
Genomic DNAs were extracted from four or five
5-μm-thick sections of formalin-fixed, paraffinembedded tissue, using a commercial kit (QIAamp
DNA Mini Kit, Qiagen, Valencia, CA). Briefly, after
Turk J Hematol 2011; 28: 15-26
deparaffinization with xylene, tissue samples were
digested with proteinase K treatment. DNAs that
emerged from cells were collected in the column
through ethanol treatment. Cellular remnants and
chemical agents were removed by washing buffer,
and genomic DNA was eluted in DNAase-free buffer
and stored at -20ºC.
Commercial kit (EZ Methylation Gold-Kit, Zymo
Research, Orange, CA) was used for bisulfite modification of isolated DNA. Briefly, DNA concentration
was measured and arranged to make 500 ng/μL, and
130 μL conversion reagent was added to 20 μL DNA
sample. Samples were incubated for 10 min at 98ºC
and then for 2.5 h at 64ºC. Samples were transferred
to column with 600 μL M-binding buffer. After homogenization, samples were centrifuged. Samples were
rinsed with 100 μL M-wash buffer, and were then
incubated in 200 μL M-desulfonation buffer for 20
min at room temperature. After incubation, samples
were rinsed two times, and then 10 μL M-elution buffer was added. Sodium bisulfite-treated genomic
DNA was stored at -20ºC.
Primer and probe sequences of the promoter
region of MGMT gene were used, which target the
localization of 1067 -1149 bp amplicon, as described
by Esteller et al. (33) (GenBank Accession Number:
X61657). Final reaction volume for analysis of both
methylated and unmethylated profiles was performed in 20 μL volume: 2 μl from each primer
(final concentration: 0.5 μM), 2 μl TaqMan probe
(final concentration: 0.2 μM), 4 μl LightCycler
TaqMan Master mixture, 5 μl DNA sample, and 5 μl
polymerase chain reaction (PCR)-grade water. The
cycling conditions for methylation-specific PCR
were: 10 min at 95ºC for Taq activation, followed by
45 cycles of 95ºC for 10 seconds (sec), 60ºC for 20
sec, and 72ºC for 1 sec for amplification. PCR products were run on a 3% agarose gel containing ethidium bromide.
Statistical Analysis
χ2 test, Mann-Whitney test and analysis of variance were applied for statistical analysis. MannWhitney test was used to analyze the relationship of
positivity of apoptosis-related proteins, PI (Ki-67)
and AI with immunophenotypic differentiation profiles (CD10/bcl6/MUM1). Spearman correlation
coefficient test was used to analyze any significant
relationship between PI, AI and percentage of posi-
Türk et al.
Apoptosis, proliferation in large B-cell lymphoma
Turk J Hematol 2011; 28: 15-26
tive cells with apoptosis-related proteins, CD10, bcl6, and MUM1, whether evaluated as positive or
negative using cut-off values. The results were considered as statistically significant when p<0.05. All
the statistics were calculated using the SPSS 11.0
program (SPSS 11.0 Inc., Chicago, IL, USA) for
Windows.
Results
Patients [53 males (52.5%), 48 females (47.5%)]
were aged between 19 and 84 (mean 55.53±14.12).
Localization was nodal in 42 (41.6%) cases andextranodal in 58 (57.4%) cases, while one case was
unknown.
Immunohistochemical expression of bcl-6, CD10,
MUM1, bcl-2, bcl-xl, bax, bak, and bid proteins was
found in 51/101 (50.5%), 20/101 (19.8%), 49/101
(48.5%), 32/101 (31.7%), 8/101 (7.9%), 67/101
(66.3%), 82/101 (81.2%), and 67/99 (66.3%) cases,
respectively (Figure 1 A-E). The mean PI was 46.15%
(±32.82) as assessed by Ki-67 staining. The mean AI
was 1.94% (±2.68) as determined by the TUNEL
method (Figure 1 F).
Two major immunophenotypic profiles were distinguished according to the pattern of differentiation
described by Hans et al. [11]: (a) GCB immunophenotypic profile: 29 cases (CD10+: 20 cases, CD10- /
bcl-6+ /MUM1-: 9 cases) and (b) non-GCB immunophenotypic profile: 72 cases (CD10- /bcl-6-: 47 cases,
CD10- /bcl-6+ /MUM1+: 25 cases) (Table 1).
Mann-Whitney test was used to analyze the association of two immunophenotypic differentiation
profiles in relation to the AI, the expression levels of
apoptosis-related proteins and PI (Ki-67) (Table 2).
Compared to the non-GCB profile, the GCB profile
was significantly associated with a higher PI
(p=0.011). However, no other significant correlations were determined between the two major differentiation immunophenotypic profiles regarding
AI and the expression levels of apoptosis-related
proteins bcl-2, bcl-xl, bax, bak, and bid (p>0.3).
The percentage of cells stained by CD10, bcl-6
and MUM1 were analyzed in relation to apoptosisrelated proteins, AI and PI by Spearman correlation
coefficient test, regardless of previous evaluation
with cut-off points to consider the results as positive
or negative (Table 3). The expression of bcl6 was
positively correlated with the expression of bcl2
19
(r=0.226, p=0.023), the AI (r=0.272, p=0.006) and
the PI (r=0.515, p<0.001), but a significantly negative correlation was observed with the expression
of bax (r=-0.221, p=0.027). The expression of
MUM1 showed significant positive correlation with
the expression of bcl-xl (r=0.295, p=0.003), bid
(r=0.313, p=0.002), AI (r=0.341, p<0.001), and PI
Table 1. CD10/bcl-6/MUM1 immunophenotypic differentiation profiles
(Total n=101)
Immunophenotypic differentiation profiles
n (%)
Germinal center B-cell-like profile
bcl-6-
CD10+
bcl-6+
bcl-6+
CD10-
MUM1-
1(1%)
MUM1+
2 (2%)
MUM1-
13 (12.8%)
MUM1+
4 (4%)
MUM1-
9 (8.9%)
TOTAL
29 (28.7%)
Non-germinal center B-cell-like profile
CD10-
bcl-6-
CD10-
bcl-6+
MUM1+
18 (17.8%)
MUM1-
29 (28.7%)
MUM1+
25 (24.8%)
TOTAL
72 (71.3%)
Table 2. The immunophenotypic differentiation profiles in relation to
the apoptotic index (AI), the expression of apoptosis related proteins,
and the proliferation index (PI) (Mann-Whitney test)
Percentage of Immunophenotypic
positive
differentiation
expression
profile
AI
bcl-2
bcl-xl
bax
bak
bid
PI (Ki-67)
Mean
rank
p
values
0.941
1.93
GCB profile
51.33
1.91
non-GCB profile
50.87
18.52
GCB profile
53.52
15.69
non-GCB profile
49.99
1.86
GCB profile
46.95
6.16
non-GCB profile
52.63
43.07
GCB profile
45.81
50.87
non-GCB profile
53.09
58.34
GCB profile
51.36
60.80
non-GCB profile
50.85
42.00
GCB profile
45.34
51.81
non-GCB profile
51.93
59.17
GCB profile
62.66
41.69
non-GCB profile
46.31
0.561
0.341
0.257
0.937
0.297
0.011*
GCB profile: Germinal center B-cell-like profile, Non-GCB profile: Non-germinal
center B-cell-like profile
*indicates the statistically significant correlations (p<0.05)
20
Türk et al.
Apoptosis, proliferation in large B-cell lymphoma
Turk J Hematol 2011; 28: 15-26
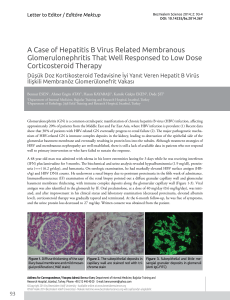
a
b
c
d
e
f
Figure 1. Immunohistochemical expression of antiapoptotic proteins (a) bcl2 and (b) bcl-xl and apoptotic proteins (c) bax, (d) bak and
(e) bid in neoplastic cells of diffuse large B-cell lymphomas (a-d, x400; e, x200). (f) Staining of apoptotic cells by the TUNEL method (x400)
(r=0.330, p=0.001). Similarly, the Spearman correlation coefficient test was used to analyze the relations
between apoptosis-related proteins, AI and PI. The
expression of bcl-xl protein showed significant posi-
tive correlation with the expression of bak (r=0.198,
p=0.047) and bid (r=0.198, p=0.049) proteins. The
expression of bax protein showed significant positive
correlation with the expression of bak (r=0.229,
Türk et al.
Apoptosis, proliferation in large B-cell lymphoma
Turk J Hematol 2011; 28: 15-26
21
Table 3. Correlations between CD10, bcl-6 and MUM1 proteins and the apoptosis related proteins, the apoptotic index (AI), and the proliferation
index (PI) (Spearman’s correlation test)
bcl-2
CD10
bcl-6
MUM1
bcl-xl
bax
bak
bid
AI
PI
r=0.092
r=0.050
r=0.055
r=-0.024
r=-0.001
r=0.130
r=0.148
p=0.360
p=0.619
p=0.585
p=0.811
p=0.991
p=0.195
p=0.139
r=0.226
r=0.176
r=-0.221
r=0.123
r=0.114
r=0.272
r=0.515
p=0.023*
p=0.078
p=0.027*
p=0.220
p=0.260
p=0.006*
p<0.001*
r=0.122
r=0.295
r=0.009
r=0.152
r=0.313
r=0.341
r=0.330
p=0.225
p=0.003*
p=0.932
p=0.129
p=0.002*
p<0.001*
p=0.001*
r, Spearman’s correlation coefficient. The positive or negative sign of the Spearman’s correlation coefficient r expresses significant (p<0.05) or nonsignificant (p=0.05 or
p>0.05) positive or negative correlations between two continuous variables
*indicates the statistically significant correlations (p<0.05)
a
Amplification Curves
Case no: 1
Case no: 2
Negative template control
2.5
2.3
Fluodescence (530)
2.1
1.9
1.7
1.5
1.3
1.1
0.9
0.7
0.5
0.3
0.1
2
4 6 8 10 12 14 16 18 20 22 24 26 28 30 32 34 36 38 40 42 44
Cycles
b
Amplification Curves
Case no: 1
Case no: 2
Negative template control
Fluodescence (530)
0.39
0.34
0.29
0.24
0.19
0.14
0.09
0.04
2
4 6 8 10 12 14 16 18 20 22 24 26 28 30 32 34 36 38 40 42 44
Cycles
Figure 2. (a) Amplification curve after real-time PCR with methyleted specific primer and probe set. (b) Amplification curve after
real-time PCR with unmethyleted specific primer and probe set
p=0.022) and bid (r=0.223, p=0.027) proteins. The
expression of bak protein also showed significant
positive correlation with the expression of bid
(r=0.214, p=0.033) protein. The AI showed significant
positive correlation with the PI (r=0.349, p<0.001).
MGMT promoter methylation analysis was performed in 95 patients with DLBCL (Figure 2). MGMT
methylated profile was found in 31/95 (32.6%) samples while MGMT unmethylated profile was determined in 64/95 (67.4%) samples. The GCB profile
was determined in 6 cases (22.2%) and non-GCB
profile in 25 cases (36.8%) out of the 31 with MGMT
methylated profile (Table 4). There was no significant association between MGMT promoter methylation status and the two immunophenotypic differentiation profiles (p=0.173) (Table 5). There was
also no significant association between MGMT promoter methylation status and the expression levels
of apoptosis-related proteins, the AI and the PI
(p>0.06).
Discussion
In the present study, we found that in DLBCLs,
the GCB profile was significantly associated with a
higher PI compared to the non-GCB profile. In addition, the percentage of cells reacting with the GC
B-cell-related bcl-6 protein showed significant positive correlation with PI. The correlation between
bcl-6 and proliferation is not conclusive in the published data. A review of the literature indicates that
bcl-6 could have a role both as stimulator or inhibitor of cell cycle progression and proliferation [16,3237]. Some in vitro studies showed that bcl-6 expression was associated with delaying cell cycle progression and decreased proliferation [15,35]. Albagli
et al. [16] demonstrated that bcl-6 mediates growth
suppression associated with impaired S phase progression in human U2OS osteosarcoma cells.
Hosokawa et al. [35] established Ba/F3 pro-B cells
carrying a human bcl-6 transgene, and revealed that
22
Türk et al.
Apoptosis, proliferation in large B-cell lymphoma
Table 4. The immunophenotypic differentiation profiles in relation to
the MGMT methylation status
MGMT methylation status
Methylated
Total number of
Unmethylated
cases
GCB
6 (22.2%)
21 (77.8%)
27
non-GCB
25 (36.8%)
43 (63.2%)
68
Total
31 (32.6%)
64 (67.4%)
95
MGMT, O6- methylguanine-DNA methyltransferase GCB profile: Germinal center
B-cell-like profile, Non-GCB profile: Non-germinal center B-cell-like profile
Table 5. Association between MGMT methylation status and expression of CD10/bcl-6/MUM1 proteins (χ2 test)
Parameter
MGMT
p
methylated (%) values*
CD10 expression
bcl-6 expression
+
5 (26.3%)
-
26 (34.2%)
+
14 (29.2%)
-
17 (36.2%)
+
12 (26.7%)
-
19 (38.0%)
CD10/bcl-
CD10+/bcl-6-/MUM1-
0
6/MUM1
CD10+/bcl-6+/MUM1-
5 (38.5%)
MUM1 expression
coexpression
CD10+/bcl-6-/MUM1+
0
CD10+/bcl-6+/MUM1+
0
CD10-/bcl-6-/MUM1+
4 (23.5%)
CD10-bcl-6-/MUM1-
13 (46.4%)
CD10-/bcl-6+/MUM1-
1 (12.5%)
CD10-/bcl-6+/MUM1+
8 (34.8%)
0.514
0.469
0.242
0.729
MGMT, O6- methylguanine-DNA methyltransferase
*indicates the statistically significant correlations (p<0.05)
induced bcl-6 protein downregulates the expression
of cyclin A2 and inhibits cell proliferation, though
Shaffer et al. [34] demonstrated that bcl-6 may
induce cell cycle progression and maintain proliferation by blocking the expression of the cyclindependent kinase inhibitor p27 and by repressing
blimp-1 expression, which decreases c-myc expression. Furthermore, Allman et al. [37] revealed that
bcl-6 protein expression was 34-fold higher in rapidly proliferating GC B-cells than in the resting
B-cells. Bai et al. [18] suggested that the resistance
to antiproliferative signals through the P19 (ARF)
-p53 pathway and downregulation of the expression
of cyclin-dependent kinase inhibitor p27 are at least
partly responsible for the association between bcl-6
and increased proliferation in DLBCL. Xu et al. [38]
investigated the role of bcl-6 gene rearrangement
Turk J Hematol 2011; 28: 15-26
and bcl-6 expression in subgroups of DLBCLs and
showed that DLBCLs with bcl-6 gene rearrangement had higher proliferative activity than those
without bcl-6 gene rearrangement. On the basis of
the previously mentioned results, results of this
study suggest that bcl-6 protein expression could be
responsible for high PI in the GCB profile of DLBCL.
Additionally, in this study, bcl-6 expression
showed significant positive correlation with the AI
and anti-apoptotic protein bcl-2 expression and
negative correlation with pro-apoptotic protein bax
expression. In association with the proliferation of
bcl-6, the data in the literature indicates that bcl-6
may have a role as stimulator or inhibitor of apoptosis [15,16,39-41]. Some studies showed that bcl-6
may protect cells from apoptosis [39,41]. Kojima et
al. [39] demonstrated that bcl-6 may have a stabilizing role to protect spermatocytes from heat shockinduced apoptosis in bcl-6-deficient mice. Baron et
al. [41] showed that the human programmed cell
death-2 (PDCD2) gene is a target of bcl-6 repression
in Epstein-Barr virus-negative Burkitt lymphoma cell
line expressing high levels of bcl-6. Furthermore,
they immunohistochemically demonstrated the
inverse relationship between bcl-6 and PDCD2
expression in human tonsils [41]. Consequently,
they proposed that bcl-6 may downregulate apoptosis by means of its repressive effects on PDCD2.
However, some other studies suggested that high
expression of bcl-6 may induce apoptosis [1416,40]. Albagli et al. [16] used the human osteosarcoma cell line U2OS transfected with bcl-6 and
demonstrated that bcl-6 mediates dose-dependent
growth suppression, which is associated with
impaired S phase progression and trigger of apoptosis. Yamochi et al. [14] showed that viability of CV-1
and HeLa cells infected with a recombinant adenovirus expressing bcl-6 was markedly reduced due to
apoptosis. Furthermore, induction of apoptosis by
bcl-6 overexpression was preceded by downregulation of apoptosis repressors bcl-2 and bcl-xl, which
suggests that bcl-6 might also regulate the expression of these apoptosis repressors [14]. Bai et al.
[18] found that high expression of bcl-6 shows significant correlation with negative bcl-2 expression.
They suggested that the association between
increased bcl-6 expression and increased apoptosis
in DLBCL might be due, at least to some extent, to
the downregulation of bcl-2, which is induced by
Turk J Hematol 2011; 28: 15-26
bcl-6 overexpression [18]. In contrast to previous
studies, in this study, we determined that bcl-6
expression shows a significantly positive correlation
with bcl-2 expression. Moreover, we found that bcl-6
expression shows significant negative correlation
with pro-apoptotic protein bax expression. It is well
known that apoptosis-related proteins bcl-2 and bclxl show anti-apoptotic effect, whereas bax, bak and
bid show pro-apoptotic function. These proteins, as
members of the bcl-2 family, exert their specific
effects by dimerizing with themselves or with each
other [25]. If the balance favors the presence of free
bcl-2, apoptosis is inhibited, whereas when bax predominates, apoptosis is initiated [25]. Thus, the ratio
of the anti-apoptotic to the pro-apoptotic proteins
determines whether a given cell will respond to or
ignore an apoptotic stimulus [25]. On the basis of
these results, we suggest that pro-apoptotic function
of bcl-6 partly occurs independent of an anti-apoptotic effect of bcl-2 and pro-apoptotic effect of bax.
In this study, no significant correlations were
determined between two major immunophenotypic
differentiation profiles regarding AI and the expression levels of apoptosis-related proteins bcl-2, bcl-xl,
bax, bak, and bid. However the percentage of cells
positive for MUM1, which is major marker of nonGCB immunophenotypic differentiation profile,
showed a significantly positive correlation with the
positivity percentage of anti-apoptotic protein bcl-xl,
pro-apoptotic protein bid, AI, and PI. Bai et al. [10]
determined that the expression of MUM1 showed
significant negative correlation with the expression
of bax and bid and significant positive correlation
with the expression of bcl-xl. They proposed that this
result may provide an explanation for the significant
positive correlation between MUM1 and bcl-xl expression in their study, because the MUM1 (IRF4) gene is
also a nuclear factor-Kappa B target. Nuclear factorKappa B, depending on the stimulus and the cellular
context, can activate pro-apoptotic (e.g. CD95,
CD95L, TRAIL receptors), anti-apoptotic (c-FLIP, bcl2, bcl-xl, c-IAP1, c-IAP2) and cell cycle (cyclin D1,
cyclin D2, c-myc) genes [32]. This status may explain
how MUM1 expression shows significant positive
correlation with pro-apoptotic protein bid, AI, PI, as
was also shown in the results of Bai et al. [10].
In this study, we determined that DLBCLs frequently express bcl-2 family member apoptosisrelated proteins as bcl-2 in 31.7%, bcl-xl in 7.9%, bax
Türk et al.
Apoptosis, proliferation in large B-cell lymphoma
23
in 66.3%, bak in 81.2%, and bid in 66.3%. Bairey et al.
[25] examined the role of bcl-2 family proteins in
aggressive NHLs and determined that of the entire
group of 44 samples, 25 (57%) showed bcl-2 staining, 11 (25%) showed bcl-xl staining, 17 (39%)
showed bax staining, and 16 (36%) showed bak
staining. Among these proteins, bcl-2, the most
exclusively investigated, is a potent suppressor of
apoptosis and is determined in unexpectedly high
levels probably in all cancers of humans [50].
Simonian et al. [43] suggested that bcl-2 and bcl-xl
can downregulate or upregulate apoptosis. Sclaifer
et al. [44] showed that all cases with a positive
expression of bax expressed either bcl-2 or mcl-1
anti-apoptotic proteins, suggesting that the presence of bax in the tumor cells must be associated
with apoptosis-inhibiting proteins, allowing malignant cell survival. Kiberu et al. [22] investigated the
correlation between apoptosis, proliferation and
bcl-2 expression in NHLs, and they suggested that
expression of bcl-2 is not necessarily related to low
levels of apoptosis, as some bcl-2 positive highgrade tumors also had high levels of apoptosis.
Nevertheless, they found that the majority of lymphomas expressing bcl-2 had average levels of
apoptosis [22]. These results suggested that bcl-2
independent apoptosis is an important factor influencing cell death in many NHLs [22]. Overall results
of previous studies [10,19,22-27] and our results
indicate that the expressions of bcl-2 family proteins
are variable and heterogeneous in DLBCL. The dual
effect of bcl-2 family proteins may show individual
variation in the pathogenesis and prognosis of
DLBCLs.
In this study, we found that the AI showed significant positive correlation with the PI. This result is
correlated with previously reported results
[10,22,45]. However, we were unable to show any
relation between apoptosis-related proteins and PI.
In this study, we determined that MGMT promoter methylation was detected in 32.6% of cases.
MGMT methylation was reported at a frequency
from 36% to 52% in Western populations in previous
studies [1,3,30,46,47]. Higher rates (75.9%) were
found in Middle Eastern populations [38]. This status may be explained by the differences in etiologic
factors such as viral infections and exposure to environmental factors as well as differences in the
genetic susceptibility. Furthermore, substantial ethnic differences existed with respect to molecular
features of malignant tumors [31,48-50].
24
Türk et al.
Apoptosis, proliferation in large B-cell lymphoma
No significant association between MGMT promoter methylation status and the two immunophenotypic differentiation profiles was observed in
this study. Al-Kuraya et al. [31] investigated the
interrelationship between MGMT methylation status and immunohistochemical coexpression of
CD10/bcl-6, and they did not show a significant
association. Furthermore, Al-Kuraya et al. determined that their frequency of GCB profile DLBCL
(13%) was somewhat lower than described in previous studies. They also found a much higher rate
of MGMT methylation in their patients (75.9%) as
compared with previous studies [1,3,30,31,46,47].
In our study, no significant association was found
between MGMT promoter methylation status and
the expression levels of apoptosis-related proteins,
the AI and the PI. There is no similar previous study
searching the relation of MGMT promoter methylation and expression levels of apoptosis-related
proteins, the AI and the PI. Further studies could
be done to elucidate the role of MGMT promote
methylation in DLBCLs.
In summary, these results suggest that bcl-6 protein expression may be responsible for the high PI
in the GCB profile of DLBCLs. Additionally, we
found that expression status of apoptosis-related
bcl-2 family proteins (bcl-2, bcl-xl, bax, bak, bid)
was not significantly associated with the immunophenotypic differentiation profiles. We also determined that there was no significant association
between MGMT promoter methylation status and
the immunophenotypic differentiation profiles of
DLBCLs.
Acknowledgements
This research project was supported by the
Turkish Society of Hematology (2007/348).
Additionally, results of this study were presented at
the 22nd European Congress of Pathology (4-9
September 2009, Florence, Italy) and the 19th
National Congress of Pathology (7-11 October 2009,
Girne, Cyprus). These presentations were supported by the Scientific Research Project Unit of
Pamukkale University. The authors thank Dr. Mehmet
Zencir for the statistical analysis.
Conflict of interest statement
None of the authors of this paper has a conflict of
interest, including specific financial interests, rela-
Turk J Hematol 2011; 28: 15-26
tionships, and/or affiliations relevant to the subject
matter or materials included.
References
1.
2.
3.
4.
5.
6.
7.
8.
9.
10.
11.
Hiraga J, Kinoshita T, Ohno T, Mori N, Ohashi H, Fukami
S, Noda A, Ichikawa A, Naoe T. Promoter hypermethylation of the DNA-repair gene O6-methylguanine-DNA
methyltransferase and p53 mutation in diffuse large
B-cell lymphoma. Int J Hematol 2006;84:248-55.
[CrossRef]
Gatter KC, Warnke RA. Diffuse large B-cell lymphoma.
In: Jaffe ES, Harris NL, Stein H, Vardiman JW, editors.
World Health Organization Classification of Tumours:
Pathology and Genetics of Tumours of Haematopoietic
and Lymphoid Tissues. Lyon: IARC Press, 2001;171-4.
Lee SM, Lee EJ, Ko YH, Lee SH, Maeng L, Kim KM.
Prognostic significance of O6-methylguanine DNA methyltransferase and p57 methylation in patients with diffuse
large B-cell lymphomas. APMIS 2009;117:87-94. [CrossRef]
Lossos IS. Molecular pathogenesis of diffuse large
B-cell lymphoma. J Clin Oncol 2005;23:6351-7.
Haralambieva E, Boerma EJ, van Imhoff GW, Rosati S,
Scuuring E, Müller-Hermelink HK, Kluin PM, Ott G.
Clinical, immunophenotypic, and genetic analysis of
adult lymphomas with morphologic features of Burkitt
lymphoma. Am J Surg Pathol 2005;29:1086-94.
Sanchez-Beato M, Sanchez-Aguilera A, Piris MA. Cell
cycle deregulation in B-cell lymphomas. Blood
2003;101:1220-35. [CrossRef]
Falini B, Mason DY. Proteins encoded by genes involved
in chromosomal alterations in lymphoma and leukaemia: clinical value of their detection by immunohistochemistry. Blood 2002;99:409-26. [CrossRef]
By the non-Hodgkin’s Lymphoma Classification
Project. A clinical evaluation of the International
Lymphoma Study Group classification of non-Hodgkin
lymphoma. Blood 1997;89:3909-18.
Küppers R, Klein U, Hansmann ML, Rajewsky K.
Cellular origin of human B-cell lymphomas. N Engl J
Med 1999;341:1520-9. [CrossRef]
Bai M, Skyrlas A, Agnantis NJ, Kamina S, Tsanou E,
Grepi C, Galani V, Kanavaros P. Diffuse large B-cell lymphomas with germinal center B-cell like differentiation
immunophenotypic profile are associated with high
apoptotic index, high expression of the proapoptotic
proteins bax, bak and bid and low expression of the
antiapoptotic preotein bcl-xl. Modern Pathol
2004;17:847-56. [CrossRef]
Hans CP, Weisenburger DD, Grenier TC, Gascoyne RD,
Delabie J, Ott G, Müller-Hermelink HK, Campo E,
Braziel RM, Jaffe ES, Pan Z, Farinha P, Smith LM, Falini
B, Banham AH, Rosenwald A, Staudt LM, Connors JM,
Armitage JO, Chan WC. Confirmation of the molecular
classification of diffuse large B-cell lymphoma by
immunohistochemistry using a tissue microarray.
Blood 2004;103:275-82. [CrossRef]
Türk et al.
Apoptosis, proliferation in large B-cell lymphoma
Turk J Hematol 2011; 28: 15-26
12.
13.
14.
15.
16.
17.
18.
19.
20.
21.
22.
23.
Colomo L, Lopez-Guillermo A, Perales M, Rives S,
Martinez A, Bosch F, Colomer D, Falini B, Montserrat E,
Campo E. Clinical impact of the differentiation profile
assessed by immunophenotyping in patients with diffuse large B-cell lymphoma. Blood 2003;101:78-84.
[CrossRef]
Shaffer AL, Rosenwald A, Hurt EM, Giltnane JM, Lam
LT, Pickarel OK, Staudt LM. Signatures of the immune
response. Immunity 2001;15:375-85. [CrossRef]
Yamochi T, Kaneita Y, Akiyama T, Mori S, Morjyama M.
Adenovirus mediated high expression of Bcl-6 in VC-1
cells induces apoptotic cell death accompanied by
down regulation of Bcl-2 and Bcl-X (L). Oncogene
1999;18:487-94. [CrossRef]
Tang TTL, Dowbenko D, Jackson A, Toney L, Lewin
DA, Dent Al, Lasky LA. The forkhead transcription factor AFX activates apoptosis by induction of the Bcl-6
transcriptional repressor. J Biol Chem 2002;277:
14255-65. [CrossRef]
Albagli O, Lantoine D, Quief S, Quignon F, Englert C,
Kerckaert JP, Montarras D, Pinset C, Lindon C.
Overexpressed Bcl-6 (LAZ3) oncoprotein triggers
apoptosis, delays S phase progression and associates
with replication foci. Oncogene 1999;18:5063-75.
[CrossRef]
Morabito F, Mangiola M, Rapezzi D, Zupo S, Oliva BM,
Ferraris AM, Spriano M, Rossi E, Stelitano C, Callea V,
Cutrona G, Ferrarini M. Expression of CD10 by B-chronic
lymphocytic leukemia cells undergoing apoptosis in
vivo and in vitro. Heamatologica 2003;88:864-73.
Bai M, Agnantis NJ, Skyrlas A, Tsangu E, Kamina S,
Galani V, Kanavaros P. Increased expression of the
bcl-6 and CD10 protein is associated with increased
apoptosis and proliferation in diffuse large B-cell lymphoma. Mod. Pathol 2003;16:471-80. [CrossRef]
Skinnider BF, Horsman DE, Dupuis B, Gascoyne RD.
Bcl-6 and bcl-2 protein expression in diffuse large
B-cell lymphoma and follicular lymphoma: correlation
with 3q27 and 18q21 chromosomal abnormalities.
Hum Pathol 1999;30:803-8. [CrossRef]
Barrans SL, Carter I, Owen GR, Davies FE, Patmore RD,
Haynes AP, Morgan GJ, Jack AS. Germinal center phenotype and bcl-2 expression combined with the
International Prognostic Index improves patient risk
stratification in diffuse large B-cell lymphoma. Blood
2002;99:1136-43. [CrossRef]
Barrans SL, Evans PA, O’Connor SJ, Kendall SJ, Owen
RG, Haynes AP, Morgan GJ, Jack AS. The t(14;18) is
associated with germinal center derived diffuse large
B-cell lymphoma and is a strong predictor of outcome.
Clin Cancer Res 2003;9:2133-9.
Kiberu SW, Pringle JH, Sobolewski S, Murphy P, Lauder
I. Correlation between apoptosis, proliferation and
bcl-2 expression in malignant non-Hodgkin’s lymphomas. J Clin Pathol (Mol Pathol) 1996;49:M268-72.
Gascoyne RD, Krajewska M, Krajewski S, Connors JM,
Redd JC. Prognostic significance of bax protein expres-
24.
25.
26.
27.
28.
29.
30.
31.
32.
33.
34.
35.
36.
25
sion in diffuse aggressive non-Hodgkin’s lymphoma.
Blood 1997;90:3173-8. [CrossRef]
Wheaton S, Netser J, Guinee D, Rahn M, Perkins S.
Bcl-2 and bax protein expression in indolent versus
aggressive B-cell non-Hodgkin’s lymphomas. Hum
Pathol 1998;29:820-5. [CrossRef]
Bairey O, Zimra Y, Shaklai M, Okon E, Rabizadeh E. Bcl2, bcl-x, bax and bak expression in short and long lived
patients with diffuse large B-cell lymphomas. Clin
Cancer Res 1999;5:2860-6.
Pagnano KB, Silva MD, Vassallo J, Aranha FJ, Saad ST.
Apoptosis regulating proteins and prognosis in diffuse
large B-cell non-Hodgkin’s lymphomas. Acta Haematol
2002;107:29-34. [CrossRef]
Sohn SK, Jung JT, Kim DH, Kim JG, Kwak EK, Park T,
Shin DG, Sohn KR, Lee KB. Prognostic significance of
bcl-2, bax, and p53 expression in diffuse large B-cell
lymphoma. Am J Hematol 2003;73:101-7. [CrossRef]
Martini M, D’Alo F, Hohaus S, Leone G, Larocca LM.
Absence of structural mutations of bak gene in B-cell
lymphoma. Haematologica 2002;87:661-2.
Dahl C, Guldberg P. DNA methylation analysis techniques. Biogerontology 2003;4:233-50. [CrossRef]
Ohno T, Hiraga J, Ohashi H, Sugisaki C, Li E, Asano H,
Ito T, Nagai H, Yamashita Y, Mori N, Kinoshita T, Naoe T.
Loss of O6-methylguanine-DNA methyltransferase protein expression is a favorable prognostic marker in
diffuse large B-cell lymphoma. Int J Hematol
2006;83:341-7. [CrossRef]
Al-Kuraya K, Narayanappa R, Siraj AK, Al-Dayel F, Ezzat
A, El Sohl H, Al-Jommah N, Sauter G, Simon R. High
frequency and strong prognostic relevance of
O6-methylguanine DNA methyltransferase silencing in
diffuse large B-cell lymphomas from the Middle East.
Hum Pathol 2006;37:742-8. [CrossRef]
Bai M, Skyrlas A, Agnantis NJ, Kamina S, Papoudou-Baj
A, Kitsoulis P, Kanavaros P. B-cell differentiation, apoptosis and proliferation in diffuse large B-cell lymphomas. Anticancer Res 2005;25:347-62.
Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman
JG. Inactivation of the DNA repair gene
O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary
human neoplasia. Cancer Res 1999;59:793-7.
Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM.
Bcl-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control.
Immunity 2000;13:199-212. [CrossRef]
Hosokawa Y, Maeda Y, Seto M. Target genes downregulated by the bcl-6/LAZ3 oncoprotein in mouse Ba/F3
cells. Biochem Biophys Res Commun 2001;283:563-8.
[CrossRef]
Shvarts A, Brummelkamp TR, Scheeren F, Koh E, Daley
GQ, Spits H, Bernards R. A senescence rescue screen
identifies BCL6 as an inhibitor of anti-proliferative
p19(ARF)-p53 signaling. Genes Dev 2002;16:681-6.
[CrossRef]
26
37.
38.
39.
40.
41.
42.
43.
44.
Türk et al.
Apoptosis, proliferation in large B-cell lymphoma
Allman D, Jain A, Dent A, Maile RR, Selvaggi T, Kehry
MR, Staudt LM. Bcl-6 expression during B-cell activation. Blood 1996;87:5257-68.
Xu FP, Liu YH, Luo XL, Zhonghua HG, Li L, Luo DL, Xu
J, Zhang F, Zhang MH, Du X, Li WY. Clinicopathologic
significance of bcl-6 gene rearrangement and expression in three molecular subgroups of diffuse large
B-cell lymphoma. Zhonghua Bing Li Xue Za Zhi
2008;37:371-6.
Kojima S, Hatano M, Okada S, Fukuda T, Toyama Y,
Yuasa S, Ito H, Tokuhisa T. Testicular germ cell apoptosis
in bcl-6-deficient mice. Development 2001;128:57-65.
Zhang H, Okada S, Hatano M, Okabe S, Tokuhisa T. A
new functional domain of bcl-6 family that recruits
histone deacetylases. Biochim Biophys Acta
2001;1540:188-200. [CrossRef]
Baron BW, Anastasi J, Thirman MJ, Furukawa Y, Fears
S, Kim DC, Simone F, Birkenbach M, Montag A, Sadhu
A, Zeleznik-Le N, McKeithan TW. The human programmed cell death-2 (PDCD2) gene is a target of bcl6
repression: implications for a role of bcl6 in the downregulation of apoptosis. Proct Natl Acad Sci U S A
2002;99:2860-5. [CrossRef]
Reed JC. Regulation of apoptosis by bcl-2 family proteins and its role in cancer and chemoresistance. Curr
Opin Oncol 1995;7:541-6. [CrossRef]
Simonian PL, Grillot DAM, Nunez G. Bcl-2 and bcl-xL
can differentially block chemotherapy induced cell
death. Blood 1997;90:1208-16.
Schlaifer D, Krajewski S, Galoin S, Rigal-Huguet F,
Laurent G, Massip P, Pris J, Delsol G, Reed JC, Brousset
P. Immunodetection of apoptosis-regulating proteins in
lymphomas from patients with and without human
immunodeficiency virus infection. Am J Pathol
1996;149:177-85.
Turk J Hematol 2011; 28: 15-26
45.
46.
47.
48.
49.
50.
Leoncini L, Cossu A, Megha T, Bellan C, Lazzi S, Luzi P,
Tosi P, Barbini P, Cevenini G, Pileri S, Giordano A, Kraft
R, Laissue JA, Cottier H. Expression of p34(cdc2) and
cyclins A and B compared to other proliferative features of non-Hodgkin’s lymphomas: a multivariate
cluster analysis. Int J Cancer 1999;83:203-9. [CrossRef]
Esteller M, Gaidona G, Goodman SN, Zagonel V,
Capello D, Botto B, Rossi D, Gloghini A, Vitolo U,
Carbone A, Baylin SB, Herman JG. Hypermethylation
of the DNA repair gene O6- methylguanine DNA methyltransferase and survival of patients with diffuse large
B-cell lymphoma. J Natl Cancer Inst 2002;94:26-32.
Rossi D, Capello D, Gloghini A, Franceschetti S, Paulli
M, Bhatia K, Saglio G, Vitolo U, Pileri SA, Esteller M,
Carbone A, Gaidano G. Aberrant promoter methylation
of multiple genes throughout the clinico-pathologic
spectrum of B-cell neoplasia. Haematologica
2004;89:154-64.
Ameyaw MM, Tayeb M, Thornton N, Forayan G, Tarig M,
Moberak A, Evans DA, Ofori-Adjei D, McLead HL.
Ethnic variation in the HER-2 codon 655 genetic polymorphism previously associated with breast cancer. J
Hum Genet 2002;47:172-5. [CrossRef]
Hartmann A, Blaszyk H, Saitoh S, Tsushima K, Tamura
Y, Cunningham JM, McGovern RM, Schroeder JJ,
Sommer SS, Koyach JS. High frequency of p53 gene
mutations in primary breast cancers in Japanese
women, a low-incidence population. Br J Cancer
1996;73:896-901. [CrossRef]
Hartmann A, Rosanelli G, Blaszyk H, Cunningham JM,
McGovern RM, Schroeder JJ, Schaid DJ, Kovach JS,
Sommer SS. Novel pattern of p53 mutation in breast cancers from Austrian women. J Clin Invest 1995;95:686-9.
[CrossRef]