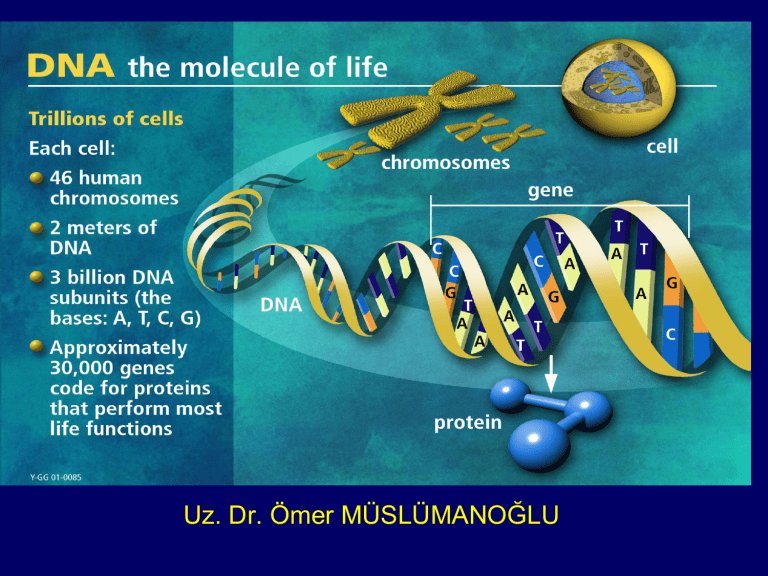
Uz. Dr. Ömer MÜSLÜMANOĞLU
DNA Yapıtaşı
Bazlar
Bazlar
Şekerler
DNA-RNA arasında temel farklar
DNA
RNA
Şeker
deoksiriboz
riboz
Baz çifti
Timin-Adenin
Sitozin-Guanin
Urasil-Adenin
Sitozin-Guanin
Yapısı
Çift sarmal
a heliks
Tek Sarmal
Düzensiz
Dayanıklılık
Stabil
DNAse ile yıkılır
Baz hidrolizine açık RNAse
ile yıkılır
Fonksyonu
Genetik bilgiyi nukleusta
saklar
Genetik bilgiyi sitoplazmaya
taşır
SANTRAL DOĞMA
replikasyon
işlenme
transkripsiyon
translasyon
DNA ISOLATION
Sources of Biological
Evidence
•
•
•
•
•
•
•
•
Blood
Semen
Saliva
Urine
Hair
Teeth
Bone
Tissue
Blood stain
Only a very small
amount of blood is
needed to obtain a
DNA profile
ORGANIC
FTA Paper
CHELEX
SDS, DTT, EDTA
and
Blood
stain
proteinase K
Blood
stain
Apply blood to
paper and allow
stain to dry
Water
INCUBATE (56 oC)
PUNCH
Centrifuge
INCUBATE (ambient)
Phenol,
chloroform,
isoamyl alcohol
Centrifuge
REMOVE supernatant
VORTEX
Centrifuge
5%
Chelex
TRANSFER aqueous (upper) phase
to new tube
REMOVE supernatant
INCUBATE (56 oC)
TE buffer
CONCENTRATE sample
(Centricon/Microcon-100 or ethanol
precipitation)
Centrifuge
QUANTITATE
DNA
WASH Multiple Times with
extraction buffer
PCR
Reagents
INCUBATE (100 oC)
Centrifuge
QUANTITATE
DNA
(NO DNA QUANTITATION
TYPICALLY PERFORMED WITH
UNIFORM SAMPLES)
PERFORM PCR
PERFORM PCR
PERFORM PCR
Figure 3.1, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
Remove a
portion of
the mixed
stain
SDS, EDTA and
proteinase K
(cell lysis buffer)
Incubate
at 37 oC
Centrifuge
Perpetrator’s sperm
mixed with victim’s
epithelial cells
sperm
pellet
SDS, EDTA and
proteinase K + DTT
“Male Fraction”
REMOVE
supernatant
DTT
lyses
sperm
heads
sperm
pellet
“Female Fraction”
Figure 3.2, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
Differential extraction used to separate sperm (male fraction) from
vaginal epithelial cells (female fraction)
female
male
male
female
Evidence (female fraction)
Evidence (male fraction)
Suspect
Victim
The four samples typically associated with a forensic DNA case…
QUANTITATION
Unknown Samples
Calibration
standards
20 ng
10 ng
5 ng
2.5 ng
1.25 ng
0.63 ng
Calibration
standards
~2.5 ng
0.63 ng
1.25 ng
2.5 ng
5 ng
10 ng
20 ng
Figure 3.3, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
Calculation of the quantity of DNA
in a cell
1. Molecular Weight of a DNA Basepair = 618g/mol
A =:
313 g/mol; T: 304 g/mol;
A-T base pairs = 617 g/mol
G = 329 g/mol; C: 289 g/mol;
G-C base pairs = 618
g/mol
2. Molecular weight of DNA = 1.85 x1012 g/mol
There are 3 billion base pairs in a haploid cell ~3 x 109 bp
(~3 x 109 bp) x (618 g/mol/bp) = 1.85 x 1012 g/mol
3. Quantity of DNA in a haploid cell = 3 picograms
1 mole = 6.02 x 1023 molecules
(1.85 x 1012 g/mol) x (1 mole/6.02 x 1023 molecules)
= 3.08 x 10-12 g = 3.08 picograms (pg)
A diploid human cell contains ~6 pg genomic DNA
4. One ng of DNA contains the DNA from 167
Importance of DNA Quantitation
(prior to multiplex PCR)
DNA amount
(log scale)
100 ng
High levels of DNA create interpretation
challenges (more artifacts to review)
-A
Too much DNA
Off-scale peaks
Split peaks (+/-A)
Locus-to-locus imbalance
+A
10 ng
2.0 ng
1 ng
Well-balanced STR multiplex
STR Kits Work Best in This Range
0.5 ng
0.1 ng
0.01 ng
Too little DNA
Heterozygote peak
imbalance
Allele drop-out
low
Locus-to-locus imbalance
Stochastic effect when amplifying
levels of DNA produces allele dropout
6 bp
deletion
X
Y
X = 106 bp
Y = 112 bp
AmpFlSTR kits
and PowerPlex 16
X = 212 bp
Y = 218 bp
PowerPlex 1.1
Female: X, X
1:1 Mixture: 3X + 1Y
Male: X, Y
Figure 5.11, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
PCR
POLİMORFİZM
• Bir popülasyonda mevcut olan genetik
çeşitliliğe polimorfizm denir
• DNA Polimorfizmi, DNA üzerinde hastalığa
neden olmayan, suskun nükleotid
değişimleri olarak tanımlanır.
(A) Sequence polymorphism
--------AGACTAGACATT--------------AGATTAGGCATT-------
(B) Length polymorphism
---------(AATG)(AATG)(AATG)---------3 repeats
---------(AATG)(AATG)---------2 repeats
Figure 2.5, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
Chromosome 12
telomere
p
(short arm)
Band 3
12p3
centromere
q
(long arm)
Band 5
12q5
telomere
Figure 2.4, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
İnsan Genomunda Dizi Tiplerinin Dağılımı
Minisatellite Marker (D1S80)
Flanking regions
Repeat region
GAGGACCACCAGGAAG
16 bp repeat unit
STR Marker (TH01)
Flanking regions
Repeat region
TCAT
4 bp repeat unit
Figure 5.1, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
Locus A
Allele 1
Allele 2
4
Homologous pair of
chromosomes
5
Allele 2
Allele 1
3
Homologous pair of
chromosomes
6
Locus B
Figure 2.6, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
1
2
3
4
5
6
5’-TTTCCC TCAT TCAT TCAT TCAT TCAT TCAT TCACCATGGA-3’
3’-AAAGGG AGTA AGTA AGTA AGTA AGTA AGTA AGTGGTACCT-5’
6
5
4
3
2
1
Figure 5.2, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
Temperature
94 oC
72 oC
72 oC
60 oC
94 oC
94 oC
94 oC
60 oC
72 oC
60 oC
Single Cycle
Time
Typically 25-35 cycles
performed during PCR
The denaturation time in the first
cycle is lengthened to ~10 minutes
when using AmpliTaq Gold to
perform a “hot-start” PCR
Figure 4.1, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
PCR
(A)
Agarose yield gel results
Smear of degraded
DNA fragments
High molecular
weight DNA in
a tight band
(B)
Degraded DNA sample
D5S818
Good quality Degraded
DNA
DNA
D13S317
D7S820
D16S539
CSF1PO
Penta D
Figure 7.1, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
(A)
miniSTRs: new tool for degraded
DNA
Smaller PCR products work better with low
Conventional
PCR primer
miniSTR
primer
copy number or fragmented DNA templates
STR repeat region
miniSTR
primer
Conventional
PCR primer
(B)
Conventional STR test
(COfiler™ kit)
150 bp smaller
MiniSTR assay (using
Butler et al. 2003 primers)
Figure 7.2, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
Y-STR
Lineage Markers
Autosomal
Y-Chromosome
Mitochondrial
(passed on in part,
from all ancestors)
(passed on complete,
but only by sons)
(passed on complete,
but only by daughters)
Figure 9.1, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
Female-Male Mixture Performance with Autosomal vs. Y-Chromosome DNA Markers
No signal observed
Female Victim
DNA Profile
Male Perpetrator
DNA Profile
DNA Profile from
Crime Scene
Autosomal STR
Profile
Y-Chromosome STR
Profile
Figure 9.2, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
Modern Use of Y-STR Testing
Captured December 13, 2003
Matching Y-STR
Haplotype Used to
Confirm Identity
(along with allele sharing
from autosomal STRs)
Uday and Qusay Hussein
Is this man really
Sadaam Hussein?
Butler, J.M. (2005) Forensic DNA Typing, 2nd Edition, Box 23.1, p. 534
Killed July 22, 2003
PCR product size (bp)
Figure A7.1, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
(A) Y-PLEX 6 (FAM-labeled loci)
PCR product size (bp)
Figure A7.2, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
ELEKTROFOREZ
Elektroforez
• Nükleik asitler (-) yüke sahitir (PO4)
• Jelde göç etmeleri büyüklükleri ve
yapıları ile ilgilidir
%
range
%
range
agarose
(kb)
acrylamide
(bp)
0.7
0.8-10
3.5
100-1000
0.9
0.5-7
5.0
80-500
1.2
0.4-6
8.0
60-400
1.5
0.2-4
12.0
40-200
2.0
0.1-3
20.0
10-100
Laser
Capillary filled with polymer
solution
(cathode)
Inlet
Buffer
5-20 kV
Detection
window
+
(anode)
Outlet
Buffer
Data
Acquisition
Sample tray
Sample tray moves automatically beneath the
cathode end of the capillary to deliver each
sample in succession
Figure 12.3, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
CAPİLLER
ELECTROPHORESİS
Cycseqpc.exe
DNA PROFİLİ
The Same 13 Locus STR Profile
in Different Populations
1 in 837 trillion
1 in 0.84 quadrillion (1015) in U.S. Caucasian population (NIST)
1 in 2.46 quadrillion (1015) in U.S. Caucasian population (FBI)*
1 in 1.86 quadrillion (1015) in Canadian Caucasian population*
1 in 16.6 quadrillion (1015) in African American population (NIST)
1 in 17.6 quadrillion (1015) in African American population (FBI)*
1 in 18.0 quadrillion (1015) in U.S. Hispanic population (NIST)
These values are for unrelated individuals
assuming no population substructure (using only p2 and 2 pq)
NIST study: Butler, J.M., et al. (2003) Allele frequencies for 15 autosomal STR loci on U.S.
Caucasian, African American, and Hispanic populations. J. Forensic Sci. 48(4):908-911.
(http://www.cstl.nist.gov/biotech/strbase/NISTpop.htm)
*http://www.csfs.ca/pplus/profiler.htm
Deciphering Artifacts from the True Alleles
Biological (PCR)
artifacts
STR alleles
Stutter products
spike
6.0%
7.8%
Dye blob
Blue channel
D3S1358
Incomplete
adenylation
+A
+A
-A
-A
D8S1179
stutter
Green channel
Pull-up
(bleed-through)
Yellow channel
Red channel
Figure 15.4, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
100%
(a)
85%
Heterozygous
peak region
>70%
MIXTURE
REGION
Stutter region
<15%
9%
100%
(b)
>70%
60%
Higher than typical
stutter product (>15%)
25%
Smaller peak area than normally seen
with heterozygote partner alleles(<70%)
10%
<15%
Wrong side of allele to be
typical stutter product
Figure 7.3, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
DNA Size (bp)
X
Y
RFUs
C
B
A
amelogenin 3 peaks at
D8S1179
X-Y peak
imbalance
B
A
C
D
4 peaks at
D21S11
A
B
CD
4 peaks at
D18S51
Figure 7.6, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
mtDNA
Human Genome
23 Pairs of Chromosomes + mtDNA
Located in cell nucleus
http://www.ncbi.nlm.nih.gov/genome/guide/
Autosomes
2 copies
per cell
Located in
mitochondria
(multiple copies
in cell cytoplasm)
mtDNA
1
2
3
4
5
6
7
8
9
10 11 12
13 14 15 16 17 18 19 20 21 22 X
Nuclear DNA
3.2 billion bp
Y
Sexchromosomes
16,569 bp
Mitochondrial
DNA
100s of copies
per cell
Figure 2.3, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
16024
16365
1
73
340
HV2
HV1
576
Control region (D-loop)
16024
OH
F 12S
rRNA
T
Heavy (H)
strand
cyt b
22 tRNAs
2 rRNAs
V
13 genes
16S
rRNA
1/16,569
P
E
ND6
L1
ND5
ND1
L2
S2
H
“16,569” bp
Light (L)
strand
ND4
OL
Q
I
M
A
N
ND2
C
Y
W
ND4L
R
ND3
9-bp
deletion
S1
COI
G
COIII
ATP6
ATP8
COII
D
K
Figure 10.1, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
DNA DİZİ ANALİZİ
Cycseqpc.exe
1
3
4
5
C
D
B
9
10
C
6
MtDNA Haplotype Groups:
1
2,3,6,8,11,13,15,16
4,9,10
5
7
12
14,17,18
7
B
11
C
B 2
A
E
12
B
15
B
8
B
13
B
F
16
14
17
B
G
18
G
G
Figure 10.2, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
Markers Used
(Biology)
High
Power of
Discrimination
(Genetics)
RFLP
Multi-Locus Probes
Multiplex STRs
RFLP
Single Locus Probes
mtDNA
Low
Slow
PolyMarker
D1S80
single STR
DQa
ABO
blood groups
Speed of Analysis
(Technology)
Fast
Figure 1.1, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
Lineage Markers
Autosomal
Y-Chromosome
Mitochondrial
(passed on in part,
from all ancestors)
(passed on complete,
but only by sons)
(passed on complete,
but only by daughters)
Figure 9.1, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
Performed separately
and preferably after
evidence is completed
Extract mtDNA
from evidence
(Q) sample
Extract mtDNA
from reference
(K) sample
PCR Amplify
HV1 and HV2 Regions
PCR Amplify
HV1 and HV2 Regions
Sequence HV1 and
HV2 Amplicons
Sequence HV1 and
HV2 Amplicons
(both strands)
(both strands)
Confirm sequence with
forward and reverse strands
Confirm sequence with
forward and reverse strands
Note differences from Anderson
(reference) sequence
Note differences from Anderson
(reference) sequence
Compare Q and K
sequences
Compare with database to
determine haplotype frequency
Figure 10.4, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
Roche (F15975)
FBI A1 (L15997)
GAAAAAGTCT TTAACTCCAC CATTAGCACC CAAAGCTAAG ATTCTAATTT AAACTATTCT
CTTTTTCAGA AATTGAGGTG GTAATCGTGG GTTTCGATTC TAAGATTAAA TTTGATAAGA
15970
15980
15990
16000
16010
16020
HV1
CTGTTCTTTC ATGGGGAAGC AGATTTGGGT ACCACCCAAG TATTGACTCA CCCATCAACA
GACAAGAAAG TACCCCTTCG TCTAAACCCA TGGTGGGTTC ATAACTGAGT GGGTAGTTGT
16030
16040
16050
16060
16070
Hypervariable Region I
16024-16365
342 bp examined
16080
Roche IA
16093 C
16126 C A 16129
ACCGCTATGT ATTTCGTACA TTACTGCCAG CCACCATGAA TATTGTACGG TACCATAAAT
TGGCGATACA TAAAGCATGT AATGACGGTC GGTGGTACTT ATAACATGCC ATGGTATTTA
16090
16100
16110
16120
16130
16140
HVI C-stretch
ACTTGACCAC CTGTAGTACA TAAAAACCCA ATCCACATCA AAACCCCCTC CCCATGCTTA
TGAACTGGTG GACATCATGT ATTTTTGGGT TAGGTGTAGT TTTGGGGGAG GGGTACGAAT
16150
16160
16170
16180
16190
16200
CAAGCAAGTA CAGCAATCAA CCCTCAACTA TCACACATCA ACTGCAACTC CAAAGCCACC
GTTCGTTCAT GTCGTTAGTT GGGAGTTGAT AGTGTGTAGT TGACGTTGAG GTTTCGGTGG
16210
16220
16230
16240
16250
16260
Roche IC
Roche IE
T
T
C
G C
CCTCACCCAC TAGGATACCA ACAAACCTAC CCACCCTTAA CAGTACATAG TACATAAAGC
GGAGTGGGTG ATCCTATGGT TGTTTGGATG GGTGGGAATT GTCATGTATC ATGTATTTCG
16270
16280
16290
16300
16310
16320
Roche ID
C
HV1
CATTTACCGT ACATAGCACA TTACAGTCAA ATCCCTTCTC GTCCCCATGG ATGACCCCCC
GTAAATGGCA TGTATCGTGT AATGTCAGTT TAGGGAAGAG CAGGGGTACC TACTGGGGGG
16330
16340
16350
16360
16370
16380
TCAGATAGGG GTCCCTTGAC CACCATCCTC CGTGAAATCA ATATCCCGCA CAAGAGTGCT
AGTCTATCCC CAGGGAACTG GTGGTAGGAG GCACTTTAGT TATAGGGCGT GTTCTCACGA
16390
16400
FBI B1 (H16391)
16410
16420
Roche (R16418)
16430
16440
SSO Probes
16093
16126
16129
16270
16278
16304
16309
16311
16362
Only 9 sites
examined
(A) mtDNA Sequences Aligned with rCRS (positions 16071-16140)
16090
16100
16110
16120
16130
16140
rCRS ACCGCTATGT ATTTCGTACA TTACTGCCAG CCACCATGAA TATTGTACGG TACCATAAAT
Q
ACCGCTATGT ATCTCGTACA TTACTGCCAG CCACCATGAA TATTGTACAG TACCATAAAT
K
ACCGCTATGT ATCTCGTACA TTACTGCCAG CCACCATGAA TATTGTACAG TACCATAAAT
(B) Reporting Format with Differences from rCRS
Sample Q
16093C
16129A
Sample K
16093C
16129A
Figure 10.8, J.M. Butler (2005) Forensic DNA Typing, 2nd Edition © 2005 Elsevier Academic Press
• İnsan genomu 3,164,700,000
nukleotidden oluşmaktadır.
• Toplam gen sayısı 29,000-36,000
arasındadır.
• Nükleotid dizilerinin %99’u bütün
insanlarda aynıdır.
• Bu güne kadar insanda 1,5 milyon kadar tek
nukleotid değişikliği bölgesi saptanmıştır.
• Tanımlanmış genlerin %50’den fazlasının
işlevleri henüz bilinmemektedir.
• Genomun yaklaşık %2’si proteinleri
kodlamaktadır.
• Proteinleri kodlamayan dizi tekrarları,
genomun büyük bölümünü oluşturur.
İnsan Genomundan Beklentilerimiz
• Moleküler Tıp
-Tanı yöntemlerinin geliştirilmesi
- Hastalıklara genetik yatkınlığın
belirlenmesi
- Genetik yapıya özgü ilaçlar
geliştirilmesi
- Gen tedavisi yöntemlerinin
geliştirilmesi
• Biyoarkeoloji, Antropoloji ve Tarih
- Değişik toplumların göç yollarının ve
akrabalıklarının araştırılması
- Y kromozom mutasyonlarının
incelenmesiyle erkek dağılımının ve
göçlerin araştırılması
• DNA Tanımlama
- Adli tıpta suçluların belirlenmesi
- Kan bağlarının saptanması
- Organ nakillerinde doku uyumunun kesin
şekilde saptanması
- Soy ağaçlarının geliştirilmesi
Kuşkular
• Genetik Bilginin Özelliği ve
Gizliliği
- Genetik bilgiye kim sahip olacak ve
kontrol edecek?
- Genetik bilgilerin gizliliği tıbbi
gizlilikten farklı mı?
• Genetik bilginin kullanılması
- Bireye ait genetik bilgilere kim
ulaşabilecek ve bu bilgileri nasıl
kullanacak?
SABRINIZ İÇİN TEŞEKKÜRLER
